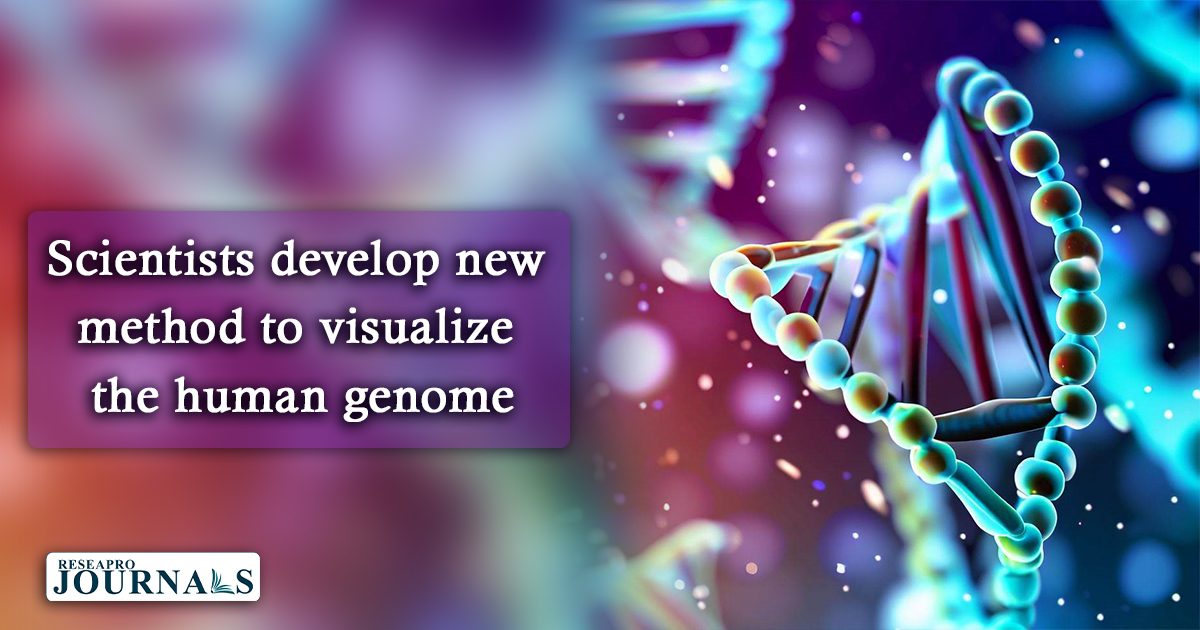
Researchers have developed a new self-supervised learning method that can improve the training of genomic models with less labeled data. The method, called Self-GenomeNet, leverages reverse-complement sequences and effectively learns short- and long-term dependencies by predicting targets of different lengths. Self-GenomeNet outperforms other self-supervised methods in data-scarce genomic tasks and outperforms standard supervised training with ~10 times fewer labeled training data. Furthermore, the learned representations generalize well to new datasets and tasks. These findings suggest that Self-GenomeNet is well suited for large-scale, unlabeled genomic datasets and could substantially improve the performance of genomic models.